Using Bacteriophage Therapy to Defeat a Resistant Bacterial Infection
Reporting in the journal Nature, clinicians describe the use of a bacteriophage to treat a flesh-eating infection by an antibiotic-resistant bacteria, with excellent clinical response. Bacteriophages, from the Greek ‘bacteria eater’, are viruses which target bacteria.
Bacteriophage (or more simply ‘phage’) therapy is being explored as a solution to the growing threat of antimicrobial resistance. Despite the exotic-sounding name, bacteriophage therapy is nothing new – in fact, its first application in 1919 predates the discovery of penicillin in 1929. However, their use has not been accompanied with robust research, meaning that there is still uncertainty regarding their use in modern medicine.
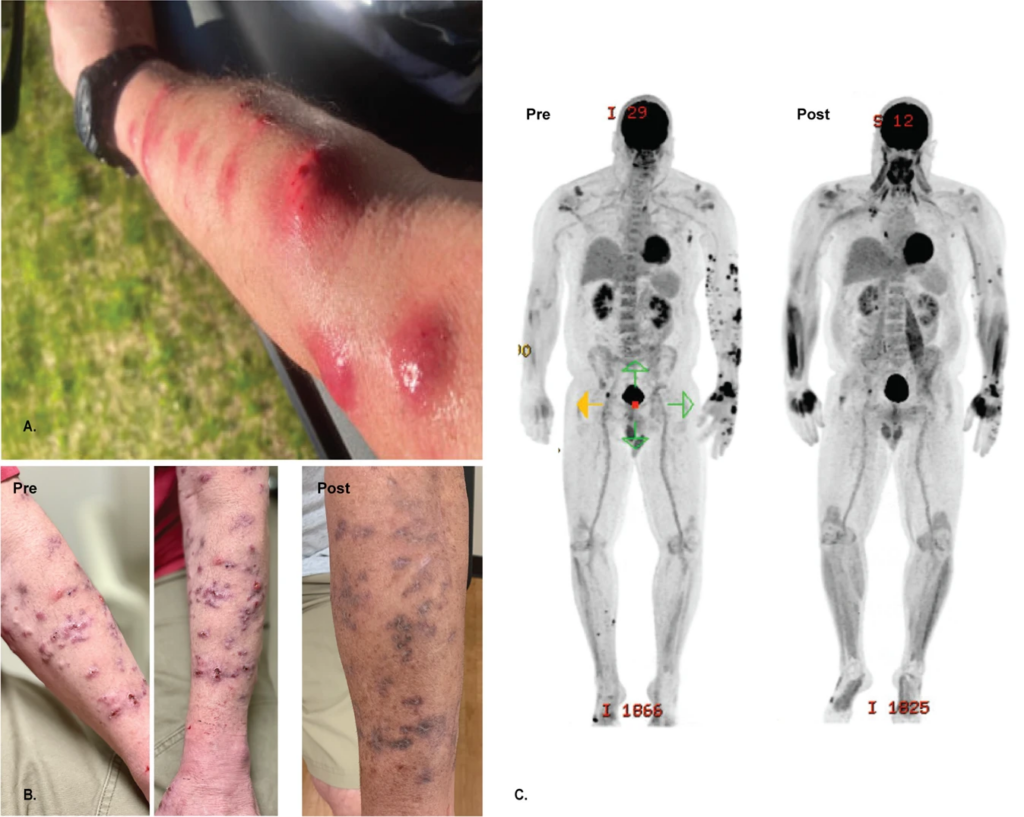
The authors report treating Mr. M, a 56 year-old man with disseminated cutaneous Mycobacterium chelonae infection with a single bacteriophage in conjunction with antibiotic and surgical management. He had previously received extensive antimicrobial courses as well as surgical debridement, but the bacterial infection persisted.
M. chelonae is a rapidly growing nontuberculous mycobacterium, ubiquitous in the environment and is known to have antimicrobial resistance. In rare cases, it causes infections in immunocompromised patients. To treat the infection, the researchers used a bacteriophage called Muddy, which had been isolated from a South African eggplant.
After the phage therapy skin started, lesions significantly improved both on examination and in PET/CT scans. Furthermore, two biopsies at two and five months post-treatment revealed no evidence of granulomas or AFB on histopathology and tissue cultures have remained negative. The patient has had no adverse events from the phage therapy and administered the intravenous therapy at home for more than six months.
Bacteriophage therapy is hampered by the development of phage resistance, which can potentially be countered using an appropriately-designed phage cocktail. In this case, the researchers were limited to Muddy, since no other phages tested were highly active against the patient’s strain of M. chelonae. Although resistance to Muddy is likely to occur, it was not detected in vitro, consistent with the infrequency of phage resistance in M. abscessus isolates. Resistance in vivo leading to loss of treatment efficacy was also not observed, which together suggest that phage resistance of NTM pathogens may not be the impediment encountered with other pathogens.
A second barrier to the successful treatment of bacterial infections with phage therapy is the complex interaction between the host immune system and the bacteriophage. In this case, the patient maintained stably improved disease and negative microbiologic and histopathologic studies despite a neutralising antibody response to the phage.
The authors suggested that the phage quickly reduced the burden of infection, allowing the ongoing antimicrobial therapy to have an effect. The phage also became self-replicating at the infection site – administration after the onset of neutralising antibodies therefore became unnecessary.
There are still significant challenges to phage therapy becoming widespread. The mains ones are 1) doctors need to know the bacterial strain behind the infection and 2) they need to have several phages on hand that specifically target that strain. Compounding the latter problem, most pharmaceutical companies are hesitant to focus on developing phage therapies. Since phage therapy is over 100 years old, it is difficult to patent and generate revenue to justify the initial development costs.